Episode 33: From probiotic mechanisms to applications, with Prof. Graciela Lorca PhD

Podcast: Play in new window | Download
Subscribe: Apple Podcasts | Spotify | RSS
This episode, we discuss how to advance from probiotic mechanisms to human applications, with Prof. Graciela Lorca PhD at the University of Florida in Gainesville, USA. Prof. Lorca talks about her experiences seeking out the mechanisms of action of a probiotic – including which molecules from bacteria may have beneficial effects – and bringing a probiotic through drug trials for use in Type 1 diabetes. They also discuss probiotic responders versus nonresponders and how dietary intake may provide clues about who will respond to an intervention.
Key topics from this episode:
- Prof. Lorca’s lab is primarily concerned with discovering the mechanisms of action of specific probiotics, including the molecules they produce that can have beneficial effects on a host.
- Knowing how a probiotic works allows scientists to select strains that are likely to be effective for a certain application.
- Prof. Lorca’s lab found that L. johnsonii produces extracellular vesicles (EVs) and that a few proteins carried in these EVs may be important markers of where and how they affect the host. She triggered antibodies against these proteins, allowing them to be tracked in the host.
- EVs are small protrusions from the bacterial membrane, and only some bacteria produce them. Evs have complex cargo, which mostly represents the metabolic state of the cell.
- Prof. Lorca studied bacteria that appeared to affect autoimmunity in animal models. In humans, administering these bacteria changed immune markers; this intervention is now in a Phase II trial with humans who have Type 1 diabetes. The bacteria may be acting in the small intestine, but they don’t colonize permanently.
- Extensive data on safety were required to advance the probiotic through to a Phase II trial. Although administering EVs could be an even safer approach, they are difficult to purify from bacteria. Prof. Lorca continues to investigate the bioactive components of these EVs to perhaps administer only those components.
- Prof. Lorca is also interested in responders versus nonresponders to a probiotic intervention. One of her clinical trials showed that people had either high lactic acid bacteria (LAB) or low LAB at baseline. For those with high levels of LAB, the levels didn’t change much over time. But for those with initially low levels of LAB, the levels increased over time. The latter responded better to treatment. Furthermore, people with high LAB were shown to consume a diet with more long-chain fatty acids, which LAB can utilize. Overall, dietary intake may be a key part of uncovering responders and nonresponders.
- Over the next ten years in this field, Prof. Lorca believes we will be able to increasingly personalize probiotics according to someone’s genetics and dietary intake. Regulatory aspects are complicated but continue to evolve.
Episode links:
- Article on how bacterial EVs may have effects on type 1 diabetes: Internalization of extracellular vesicles from Lactobacillus johnsonii N6.2 elicit an RNA sensory response in human pancreatic cell lines
- Tracking bacterial EVs in the host: Identification of Biomarkers for Systemic Distribution of Nanovesicles From Lactobacillus johnsonii N6.2
- Clinical trial showing feasibility of using a probiotic strain in applications related to type 1 diabetes: Lactobacillus johnsonii N6.2 Modulates the Host Immune Responses: A Double-Blind, Randomized Trial in Healthy Adults
- Using diet to predict levels of bacteria in the gut: Identification of food and nutrient components as predictors of Lactobacillus colonization
- Master of Science degree at University of Florida: Microbiome in Health and Disease
Additional resources:
About Prof. Graciela Lorca PhD:
Dr. Graciela Lorca is currently a Professor in the Department of Microbiology and Cell Science at the University of Florida. She completed her Licentiate in Genetics studies at the National University of Misiones and later received her doctoral degree in Food Technology at the National University of Tucuman in Argentina. She completed her postdoctoral studies at the University of California San Diego in Molecular Microbiology and at the University of Toronto in Structural Biology and Gene Regulation. Since joining the Department of Microbiology and Cell Science at the University of Florida in 2007, Dr. Lorca has focused on the identification of environmental signals that modulate host-microbe interactions. Using multiomic approaches, her laboratory is investigating the bacterial components such as extracellular vesicles that target host pathways involved on those beneficial interactions in vitro and in vivo. Furthermore, Dr. Lorca’s laboratory is currently conducting human trials to evaluate the use of Lactobacillus johnsonii Type 1 Diabetes patients. Dr. Lorca currently teaches a graduate and undergraduate level Probiotics course. She is also in charge of the new concentration on Microbiome in health and disease within the Online Master program at Department of Microbiology and Cell Science.






























 Giving a lecture on beneficial microbes is hard enough to peers sitting in the back of the room, but to do so with young South Africans was more somewhat daunting. However, it proved to be a lot of fun especially when we had to identify kids who were good leaders (the boys all pointed to a girl), who liked to make stuff and sell it to others (two boys stood out). By the end, we had picked the ‘staff’ of a new company.
Giving a lecture on beneficial microbes is hard enough to peers sitting in the back of the room, but to do so with young South Africans was more somewhat daunting. However, it proved to be a lot of fun especially when we had to identify kids who were good leaders (the boys all pointed to a girl), who liked to make stuff and sell it to others (two boys stood out). By the end, we had picked the ‘staff’ of a new company.
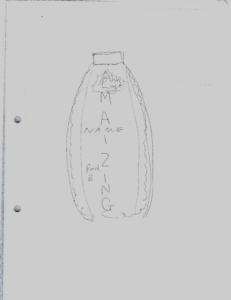 When we re-assembled to present the results, I was impressed with what could be created in such a short time. My favourite was the Amazing Maize, a bottle shaped like a corn cob with the idea it would contain fermented maize. It emphasized the importance of marketing and for products to taste and look good to be purchased.
When we re-assembled to present the results, I was impressed with what could be created in such a short time. My favourite was the Amazing Maize, a bottle shaped like a corn cob with the idea it would contain fermented maize. It emphasized the importance of marketing and for products to taste and look good to be purchased.