Archive Highlight: Microbes that break down mucus and milk to benefit the host, with Dr. Clara Belzer PhD
Podcast: Play in new window | Download
Subscribe: Apple Podcasts | Spotify | RSS
We discuss microbes, mucus, and milk with Dr. Clara Belzer PhD from Wageningen University in the Netherlands in this episode. Dr. Belzer, a molecular geneticist, specializes in studying the microorganisms that are equipped to break down the glycans in mucus and human milk within the host environment.
Key topics from this episode:
- Dr. Belzer’s research focuses on the microbes living in the host that survive on glycans (chains of sugars) produced by the host: milk oligosaccharides and mucus. The host is not good at digesting these sugars, but can use them when they’re separated into smaller components. These long chains of sugars end up in the large intestine, where certain microbes begin to digest them.
- There seems to be an evolutionary adaptation that sustains the symbiotic relationship between human milk and bacteria in the infant gut; many immune molecules in the human milk suppress pathogens, so the human milk oligosaccharides (HMOs) are available to the bacteria in the infant gut that can break them down. The bacteria are not suppressed by the acidic environment in the infant gut.
- Human milk is the best food for infants, but innovations in infant formula may make it more similar to human milk.
- Akkermansia is a genus of bacteria mostly found in adults, but also sometimes in infants, which grows in the mucosal layer of the intestines. (It doesn’t survive on dietary glycans.) Dr. Belzer’s hypothesis is that the environment created by human milk in the infant gut also fosters bacteria that can grow on mucus, creating a succession of host-benefitting bacteria. They found that HMOs, in addition to mucus, can support the growth and survival of Akkermansia, potentially helping it build a microbial network.
- There’s a genetic component to the HMOs contained in human milk; similarly, the sugar content in the mucosal glycans is related to host genetics.
- Lean individuals have a higher abundance of Akkermansia; these bacteria improve metabolism (for example, increasing insulin sensitivity) and have effects on the immune system, which both contribute to a lean phenotype. The root of these effects may be the strengthening of the gut barrier, which dampens signals from the lumen.
- Dr. Belzer has used both omics and culture-based approaches in her research. As part of her research she tries to make microbial synthetic communities, growing them in the lab and stimulating them with different glycans. This technique yields insights about the functions and microbial ecology in the gut.
- Killed Akkermansia are still able to bring health benefits to the host. Dr. Belzer had the idea that the pili structures on the bacteria were what communicated with the host, and sure enough, this was borne out in a study that showed the proteins in the pili (Amuc_1100) remained intact in the pasteurized bacteria and could stimulate the host immune system. This is a valuable finding because Akkermansia are difficult to culture.
- When Akkermansia fails to occupy the niche in the mucus layer, Bacteroides species may occupy the niche instead, forming a different microbial community in the mucus. Research is ongoing about the effects of different microbes carrying out similar functions for the host. Furthermore, scientists have many more microbial functions to discover.
Episode abbreviations and links:
- Paper showing the growth of Akkermansia on HMOs: Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro
- Study showing attenuation of diet-induced obesity in mice with Akkermansia: Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity
- Study showing the Amuc_1100 protein mechanism for Akkermansia: A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice
About Dr. Clara Belzer PhD:
Dr. Clara Belzer is Associate Professor Microbiology at the Laboratory of Microbiology of Wageningen University. The Belzer group is called ‘Microbes Mucus and Milk’ and the research is focused on the interaction of the gut microbiome with host mucus and milk. After obtaining her PhD at the Erasmus Medical Center Dr. Belzer did a postdoc at Harvard medical school. By now Dr. Belzer has years of experience on gut microbiome studies on anaerobes, including synthetic communities and different biotic concepts, with a special interest for the Akkermansia muciniphila. The group of Dr. Belzer works on several microbiome HMO and mucus related topics funded by national and international grants, some also in collaboration with medical centers and industry.
Sign up for our monthly newsletter
Follow us on LinkedIn, Bluesky, X, Facebook,










































 Giving a lecture on beneficial microbes is hard enough to peers sitting in the back of the room, but to do so with young South Africans was more somewhat daunting. However, it proved to be a lot of fun especially when we had to identify kids who were good leaders (the boys all pointed to a girl), who liked to make stuff and sell it to others (two boys stood out). By the end, we had picked the ‘staff’ of a new company.
Giving a lecture on beneficial microbes is hard enough to peers sitting in the back of the room, but to do so with young South Africans was more somewhat daunting. However, it proved to be a lot of fun especially when we had to identify kids who were good leaders (the boys all pointed to a girl), who liked to make stuff and sell it to others (two boys stood out). By the end, we had picked the ‘staff’ of a new company.
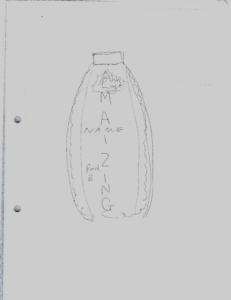 When we re-assembled to present the results, I was impressed with what could be created in such a short time. My favourite was the Amazing Maize, a bottle shaped like a corn cob with the idea it would contain fermented maize. It emphasized the importance of marketing and for products to taste and look good to be purchased.
When we re-assembled to present the results, I was impressed with what could be created in such a short time. My favourite was the Amazing Maize, a bottle shaped like a corn cob with the idea it would contain fermented maize. It emphasized the importance of marketing and for products to taste and look good to be purchased.